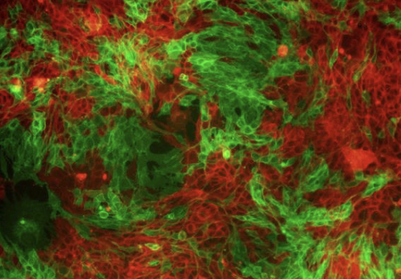
Tumor Heterogeneity, Epithelial to Mesenchymal Transition (EMT), and Metastasis. We study the heterogeneity that arises in tumors when a subset of cells re-express developmental transcription factors (TFs) and undergo an EMT or gain stem/progenitor cell characteristics. We are using biochemical and molecular approaches (mass spectrom-etry, RNAs-seq, and ChiP-seq), coupled with in vivo approaches to understand how crosstalk between tumor cells leads to enhanced metastasis, and how TFs regulate metastasis cell autonomously and non-cell autonomously.
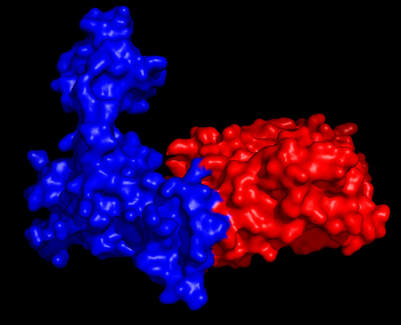
Targeting Developmental Transcription Factors to Inhibit Tumor Progression. In collaboration with the Zhao laboratory in the Department of Biochemistry and Molecular Genetics, we have identified novel small molecules that can target the Six/Eya interface or the Eya Tyr phosphatase activity in an attempt to develop targeted therapies directed at metastasis. The small molecules are currently being tested in cell based and animal models, and are anticipated to inhibit tumor growth and metastases while conferring limited side effects due to the paucity of expression of these developmental TFs in differentiated cells. This project utilizes biophysical, biochemical, and structural approaches, as well as cell-based/animal model approaches.

Understanding how cancer cells evade the immune microenvironment. Our laboratory is studying how the Eya Thr phosphatase acts, and how, when expressed in cancer cells, it contributes to the ability of the cells to avoid attack by the immune system. Recent studies have focused on an Eya-Myc-PD-L1 axis that suppresses CD8+ cytotoxic T-cells, and ongoing studies examine other immune populations as well as how EMT contributes to tumor immune evasion.
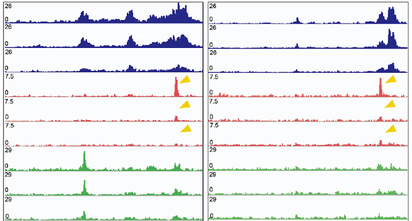
Understanding the role of the Six/Eya transcriptional complex in pediatric tumors. Using zebrafish (in collaboration with The Artinger Lab from Craniofacial Biology) as well as mouse models, we are studying how developmental transcription factors (TF) such as Six and Eya are regulated during normal embryogenesis (with a focus on microRNA mediated control of developmental TFs), as well as how their dysregulation contributes to tumor progression in various pediatric malignancies including rhabdomyosarcoma, medulloblastoma, and Ewing sarcoma
Proudly powered by Weebly



